Research
Cardiovascular disease is the leading cause of death in the industrialized world. My research vision is to integrate computational modeling, experiments and clinical data to better understand the heart’s function and structure in order to predict its behavior. With this multidisciplinary approach, we plan to create innovative translational tools that bridge the gap between basic science and clinical practice. Such computational tools have the potential to replace invasive diagnostic exams, improve their accuracy and better inform physicians about treatment efficacy. However, there are several challenges that arise when mechanistic computer models are linked to data from the clinical setting, which is tipically noise and sparse. Additionaly, the high computational cost of sophisticated computer models can pose a challenge to translate these tools into the clinical practice. For these reasons, we use and develop a number of tools to integrate complex models with sparse and noisy data to make meaningful predictions. Currently, we focus on the following areas:
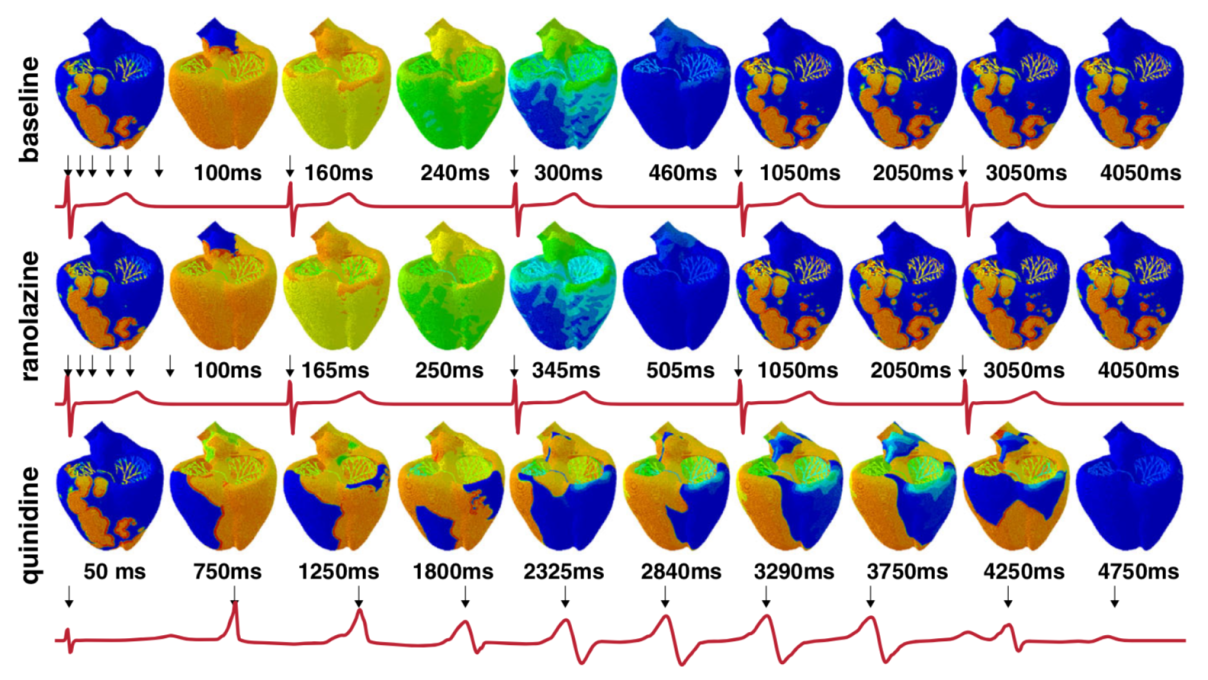
Drug-induced arrhythmias
Any drug can bind to the ion channels of cardiac cells and affect their electrical behavior. Determining the cardiac safety of a new drug is key step before it can be delivered to patients. In this line, we have developed multi-scale models that can trace the effects of a drug from the sub-cellular scales to the entire heart. However, these models are computationally demanding and have paired them with machine learning tools to quickly make predictions and explore the uncertanties in the data and the sensitivies of the model.
Selected publications:
- F. Sahli, J. Yao, E. Kuhl. Predicting drug-induced arrhythmias by multiscale modeling. International journal for numerical methods in biomedical engineering 34, no. 5 (2018): e2964.
- F. Sahli, K. Seo, E. Ashley, E. Kuhl. Classifying drugs by their arrhythmogenic risk using machine learning. bioRxiv. doi:10.1101/545863.
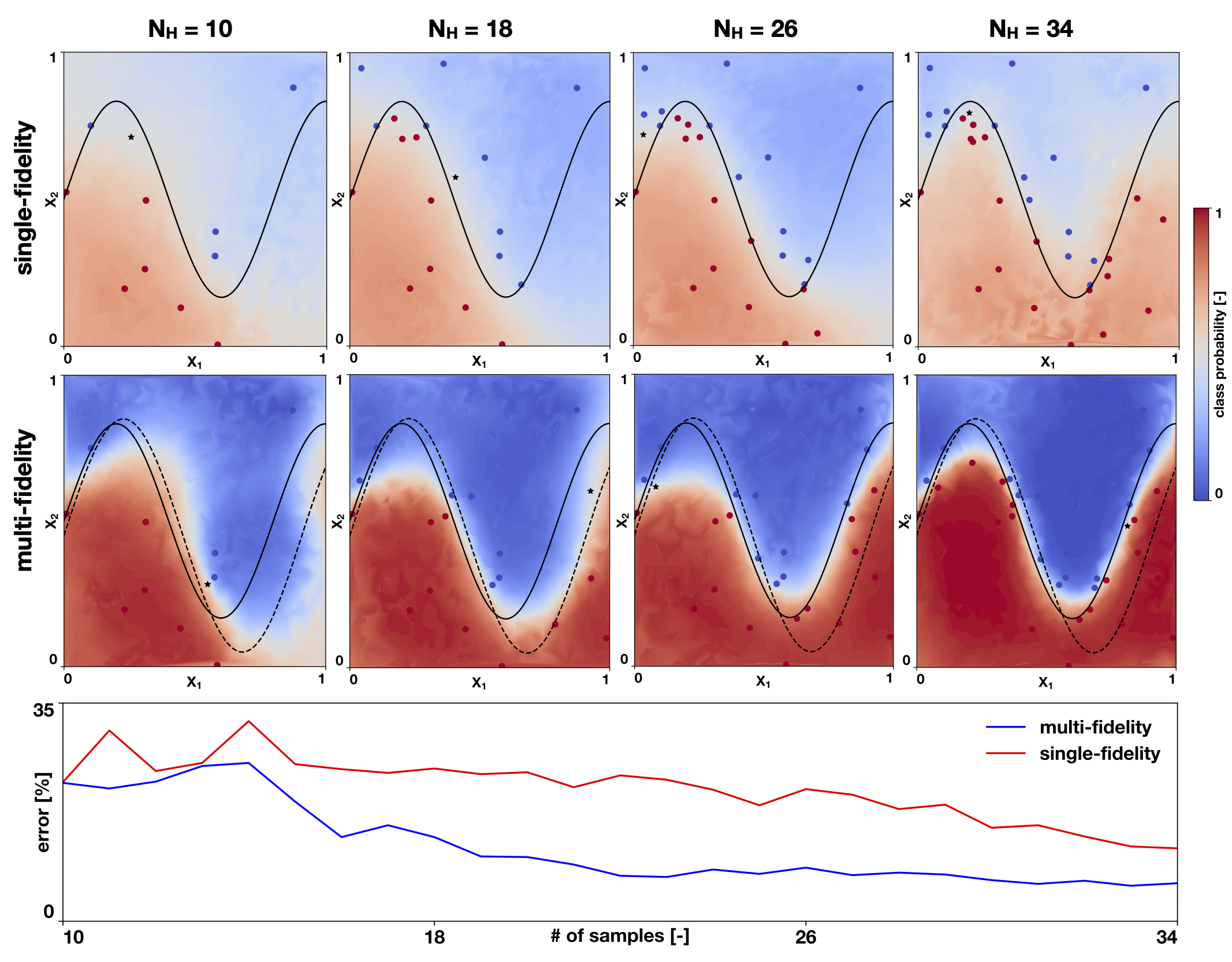
Machine learning for computational models
The high computational cost of detailed mechanistic cardiac models hinders our ability to extract knowledge from the model. To tackle this problem, we develop machine learning tools that advantage of simplified and computationally inexpensive models to precisely predict the output of the detailed models. These methods are usually called ‘multi-fidelity’, since we represent the underlying physics with different levels of detail. We have used and developed these methods in the context of cardiac electrophysiology, but they are applicable to a larger set of problems.
Selected publications:
- F. Sahli, K. Matsuno, J. Yao, P. Perdikaris, E. Kuhl. Machine learning in drug development: Characterizing the effect of 30 drugs on the QT interval using Gaussian process regression, sensitivity analysis, and uncertainty quantification. Computer Methods in Applied Mechanics and Engineering, no. 348 (2019): 313-333
- F. Sahli, P. Perdikaris, E. Kuhl, DE. Hurtado. Multi-fidelity classification using Gaussian processes: accelerating the prediction of large-scale computational models. Computer Methods in Applied Mechanics and Engineering, accepted (2019).
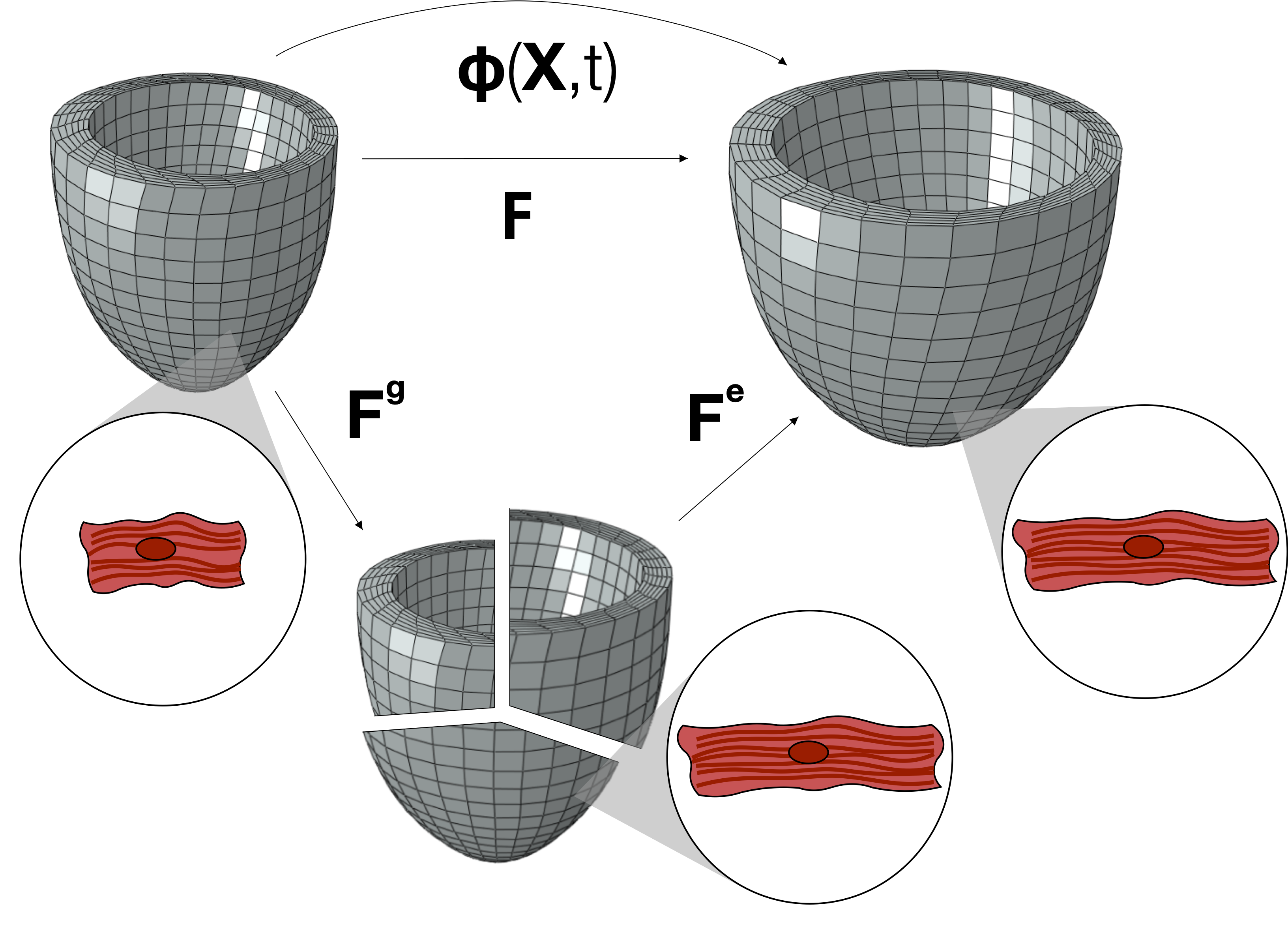
Growth and remodeling in the heart
The heart is constantly adapting its shape and properties to respond to changing demands. We have developed models to simulate the response of the heart in the presence of volume overload. We use an experimental dataset that links the cellular scales to the organ level, which is noisy and sparse, to calibrate our model. We use Bayesian statistics to compare the uncertainty in the data to the uncertainty our models. The models that we have developed are applicable to many cardiac diseases and the methodology that we have developed to calibrate the model is applicable to other problems where data is scarse.
Selected publications:
- F. Sahli, JS. Choy, KL. Sack, JM. Guccione, G. Kassab, E. Kuhl. Multiscale characterization of heart failure. Acta Biomaterialia, no. 86 (2019): 66-76
- M. Peirlinck, F, Sahli, KL. Sack, JS. Choy, GS. Kassab, JM. Guccione, M. De Beule, P. Segers, E. Kuhl. Using machine learning to characterize heart failure across the scales. Biomechanics and Modeling in Mechanobiology. 2019; doi:10.1007/s10237-019-01190-w.